At the PiE 2023 conference in Crete, Greece, Dr. Emlyn Davies from SINTEF, Norway, presented PyOPIA: Python Ocean Particle Image Analysis toolbox.
PyOPIA aims to provide a pipeline-based workflow as a standard for analysis of particle images in the ocean. The pipeline should be consistent across different instruments (hardware), and PyOPIA therefore has flexibility to adapt analysis steps to meet instrument-specific processing needs (i.e., holographic reconstruction). PyOPIA is hardware agnostic and can be used with *any* hardware used for capturing images of oceanic particles: LISST-Holo2, UVP, IFCB, SilCam, FlowCam, PlanktoScope, CytoSub, LOPC, iCPRholo etc. etc.
While all these systems record digital images, they are in different formats (colors or grayscale; resolution; noise types/sources), subject to different processing algorithms by different software prior to the extraction of the actual particle characteristics that may then be further aggregated and analyzed. This software diversity may lead to challenges in combining outputs from different systems. Such challenges may range from simple differences in output data format and how particle data are presented, to fundamental discrepancies in how accurately complex particles are segmented and how specific particle metrics (such as size) are calculated. The exact image processing steps used by different software are not always well documented and logged as metadata, making it difficult to reproduce results. These differences make the incorporation of data from the disparate instruments cumbersome or impossible. Finally, emerging challenges in the modern age of robotic adaptive sampling require particle image analysis software capable of rapid integration with robotics control system software with the ability of running in real-time on offline (or edge) computers.
The PyOPIA toolbox aims to solve these issues and has been developed by Dr. Davies over several years in collaboration with University of Plymouth, NTNU, The MBA, George Washington University, and National Oceanography Center (Southampton). It is open-source and constantly under development; new contributors are welcome.
As examples of what PyOPIA can do, Dr. Davies presented several examples:
PyOPIA will count particles from various instruments into size bins in the same way, regardless of instrument. This makes combining data from multiple, different types of instruments very easy, as the conversion from multiple units (e.g., 1/l or 1/l/um to a common unit (e.g., ul/l) for merging size spectra is handled automatically by PyOPIA:
In the example above, data from a LISST-100X and two imaging systems are automatically binned to LISST-100X size classes and subsequently merged to create the full-size spectrum.
PyOPIA can also make depth-integrated particle montages.:
In the example above, all particles from a 300 m deep cast with a SilCam and an UVP-6 are combined into two montages showing ALL the particles imaged by each instrument during the cast, as well as the particle volume distribution profiles from each instrument for immediate comparison. It is also possible to pinpoint locations of each unique particle, where, for example the two copepods expanded are from a denser region of zooplankton in the surface region indicated by the blue ovals.
PyOPIA will also generate the size distribution from a montage: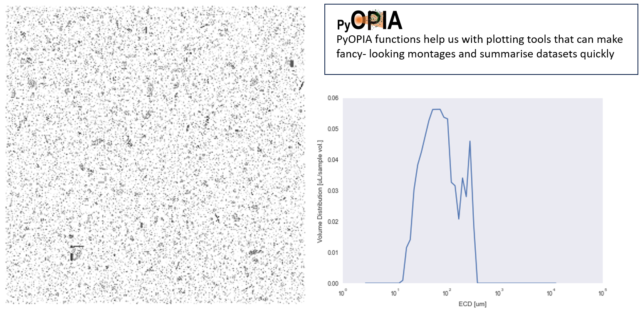
In this last example, the montage (above, left) shows all particles (23,345) imaged in a 1 nautical mile segment of an iCPRHolo tow. Also shown is the size distribution for all particles; a prevalence of small flocs < ~100 um is apparent.
It is up to the user how they want to combine and show the data; PyOPIA provides toolboxes for almost any conceivable way of organizing and plotting your data. At the same time, metadata are included, enabling accurate reproduction of the data later on if desired.
PyOPIA represents the start of a standardised open framework for particle image processing in the ocean community. For development to continue, wider community adoption, feedback and input is needed. If you are interested in trying out PyOPIA, all necessary information can be found here: https://pyopia.readthedocs.io/
PyOPIA Feedback can be provided directly to Dr. Davies.

